Case study: RNA-sequencing of liver tissue from double-crested cormorant (DCCO) embryos
This short tutorial below demonstrates how to run Seq2Fun. We use a RNA-seq dataset from a real non-model organism double-crested cormorant (DCCO), treated with ethinyl estradiol (EE2) as a show case.
1. Description of experiment
DCCO embryos were exposed via egg injection to EE2, a synthetic estrogen that is the active substance in some forms of birth control. Livers were harvested after 14 days exposure and immediately frozen in liquid nitrogen for total RNA extraction. Total RNA was sent to Genome Quebec (Montreal, Quebec, Canada), to build sequencing library with TruSeq RNA Library Prep Kit (San Diego, California, United States) before submitted to Illumina NovaSeq 6000 (San Diego, California, United States) for 100 bp PE reads sequencing.
2. Experimental samples
Each sample was subsampled with 5 million reads, just for demonstration purpose.
The samples can be download from here
| Group | Chemicals (dose) | Number of samples | Number of reads |
|---|---|---|---|
| High | EE2 (31.9 mg/ml) | 5 | 25,000,000 |
| Medium | EE2 (2.3 mg/ml) | 5 | 25,000,000 |
| Control | DMSO | 5 | 25,000,000 |
3. Database used
| Group | Number of proteins | Number of Orthologs | Number of species | Database name |
|---|---|---|---|---|
| Birds | 482,205 | 24,076 | 31 | birds.tar.gz |
1. Preparing sample.txt file
This file consists of 4 columns and must be separated by tab '\t'. The first column is the prefix name of each sample, and the second is the forward reads file, the last column is the reverse reads file. If you have a single-end (SE) reads, remove the reverse reads (second) column.
A1.CE2-S1 A1.CE2-S1_R1.fastq.gz A1.CE2-S1_R1.fastq.gz control
A3.CE1-M2 A3.CE1-M2_R1.fastq.gz A3.CE1-M2_R1.fastq.gz middle
A4.CE1-H5 A4.CE1-H5_R1.fastq.gz A4.CE1-H5_R1.fastq.gz high
B1.CE2-S2 B1.CE2-S2_R1.fastq.gz B1.CE2-S2_R1.fastq.gz control
B3.CE1-M3 B3.CE1-M3_R1.fastq.gz B3.CE1-M3_R1.fastq.gz middle
C1.CE2-S3 C1.CE2-S3_R1.fastq.gz C1.CE2-S3_R1.fastq.gz control
C3.CE1-M4 C3.CE1-M4_R1.fastq.gz C3.CE1-M4_R1.fastq.gz middle
D1.CE2-S4 D1.CE2-S4_R1.fastq.gz D1.CE2-S4_R1.fastq.gz control
D3.CE1-M5 D3.CE1-M5_R1.fastq.gz D3.CE1-M5_R1.fastq.gz middle
E1.CE2-S5 E1.CE2-S5_R1.fastq.gz E1.CE2-S5_R1.fastq.gz control
E3.CE1-H1 E3.CE1-H1_R1.fastq.gz E3.CE1-H1_R1.fastq.gz high
F3.CE1-H2 F3.CE1-H2_R1.fastq.gz F3.CE1-H2_R1.fastq.gz high
G3.CE1-H3 G3.CE1-H3_R1.fastq.gz G3.CE1-H3_R1.fastq.gz high
H2.CE1-M1 H2.CE1-M1_R1.fastq.gz H2.CE1-M1_R1.fastq.gz middle
H3.CE1-H4 H3.CE1-H4_R1.fastq.gz H3.CE1-H4_R1.fastq.gz high
2. Running Seq2Fun to quantify RNA-seq reads.
Using profiling mode to produces 3 main tables
1). s2f_id/ortholog abundance table for all samples,
2). s2f_id/ortholog abundance table for submitting to expressanalyst,
3). s2f_id/ortholog annotation table,
4). a html report summarizing all samples.
S2F_HOME/bin/seq2fun --sampletable sample.txt --tfmi S2F_HOME/database/birds/birds.fmi --genemap S2F_HOME/database/birds/birds_annotation.txt -w 8 --profiling -V --outputMappedCleanReads --outputReadsAnnoMap
1.1 An important html report to summarize all samples(please check it first) (Seq2Fun_summary_all_samples.html).
1.2 s2f id abundance for all the samples table (S2fid_abundance_table_all_samples.txt).
This table has s2f id, sample names and annotation separated by '\t'. (how many reads have assigned to the s2f id), the full name of the assigned s2f id annotation.
#NAME A1.CE2-S1 A3.CE1-M2 A4.CE1-H5 B1.CE2-S2 B3.CE1-M3 C1.CE2-S3 C3.CE1-M4 D1.CE2-S4 D3.CE1-M5 E1.CE2-S5 E3.CE1-H1 F3.CE1-H2 G3.CE1-H3 H2.CE1-M1 H3.CE1-H4 annotation2. The following tables and figures are from (Seq2Fun_summary_all_samples.html).
#CLASS:XX control middle high control middle control middle control middle control high high high middle high -
s2f_7617 158 338 475 130 329 178 342 185 479 160 312 429 241 309 345 K04198|GO:0004930;GO:0004962;GO:0005886;GO:0007165;GO:0007186;GO:0008217;GO:0016020;GO:0016021;GO:0042310;GO:0048484;GO:0086100|EDNRB|endothelin receptor type B
s2f_7570 10 9 9 8 1 8 23 8 23 9 9 7 15 7 12 K07437|GO:0004497;GO:0005506;GO:0016491;GO:0016705;GO:0020037;GO:0046872|CYP26A1|cytochrome P450 26A1
s2f_14150 0 1 0 0 0 0 0 0 0 1 1 0 1 0 0 K05254|GO:0005179;GO:0005576;GO:0007165;GO:0016608|GHRL|appetite-regulating hormone
s2f_6367 22 43 48 50 50 31 55 29 38 16 52 45 40 40 45 K03370|GO:0005794;GO:0006486;GO:0008373;GO:0016020;GO:0016021;GO:0016740;GO:0016757;GO:0097503|ST3GAL5|lactosylceramide alpha-2,3-sialyltransferase isoform X1
s2f_7077 42 81 44 63 61 61 63 66 60 83 46 65 72 74 51 U|GO:0000775;GO:0000776;GO:0005694|CFDP1|craniofacial development protein 1
s2f_12441 3 4 8 4 2 1 3 0 13 2 3 7 6 4 5 K15680|GO:0016491|HSD11B1L|hydroxysteroid 11-beta-dehydrogenase 1-like protein isoform X1
...
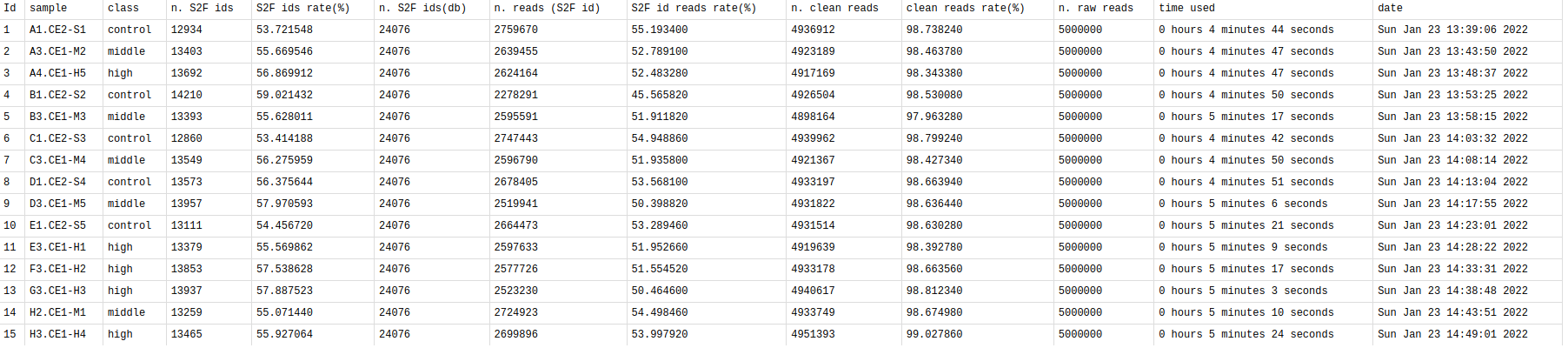 This table summarizes the general information of your samples.
This table summarizes the general information of your samples.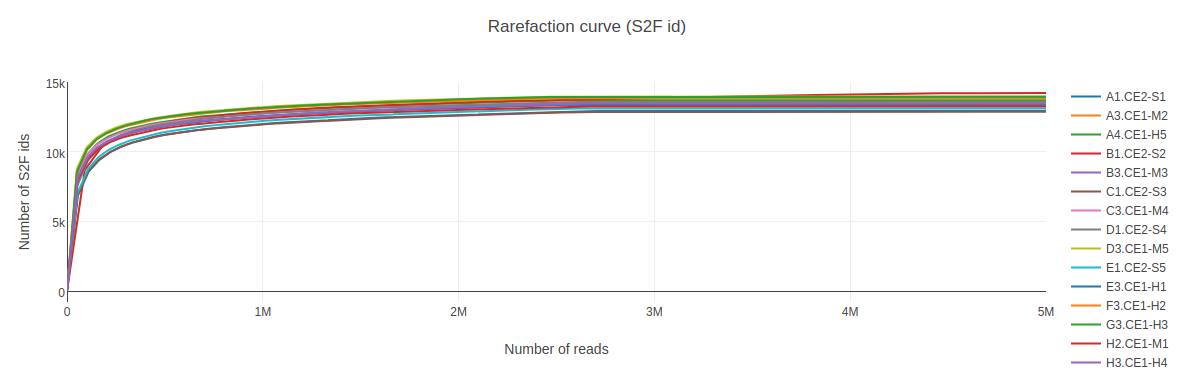 Rarefaction curve (2d) for all samples.
Rarefaction curve (2d) for all samples.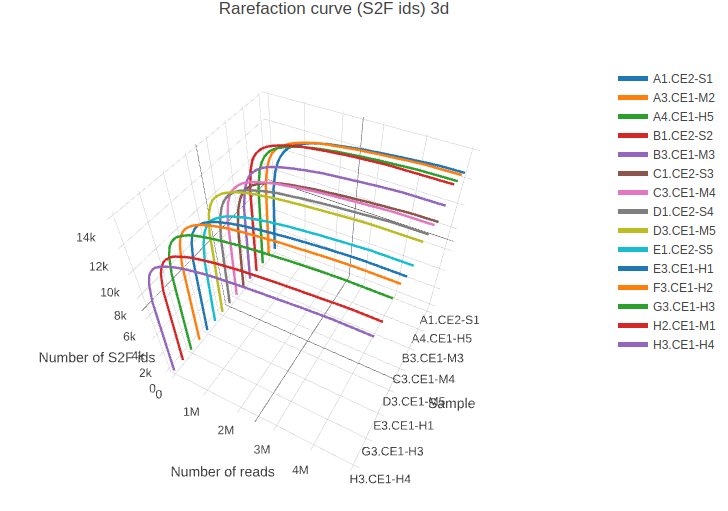 Rarefaction curve (3d) for all samples.
Rarefaction curve (3d) for all samples.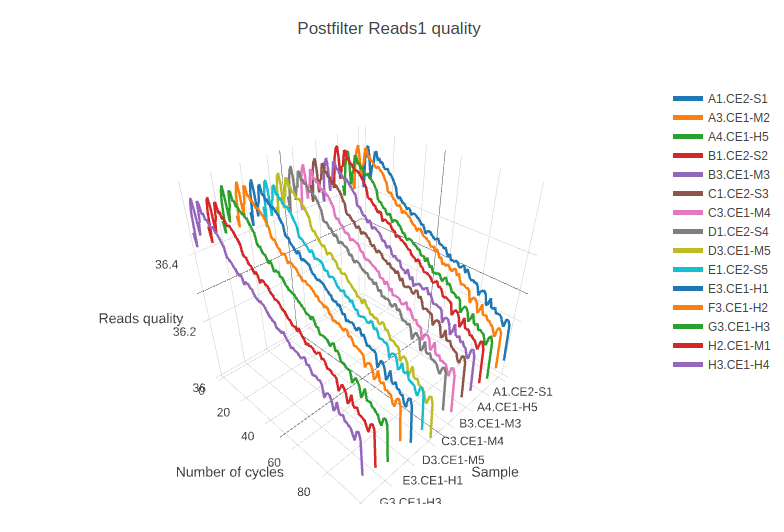 Reads quality (3d) for all samples.
Reads quality (3d) for all samples.
1. Online website-based analysis tool ExpressAnalyst
S2fid_abundance_table_all_samples_submit_2_expressanalyst.txt, this will be used to submit to ExpressAnalyst.
2. Based on the KO abundance table generated by Seq2Fun (version 1, from verion 2, we provide the s2f id abundance table), differentially expressed KOs are
identified by limma (R package), and enriched pathways are analysized (GSEA) by HTSanalyzeR
(R package).
We provide the R script (click here to download) to conduct pathway enrichment.
Please note that the following figure is generated by HTSanalyzeR, which may no longer be installed on current R version.
Therefore, we have developed an alternative R script using fgsea for pathway analysis and we suggest users to try the casestudy_fgsea.R.
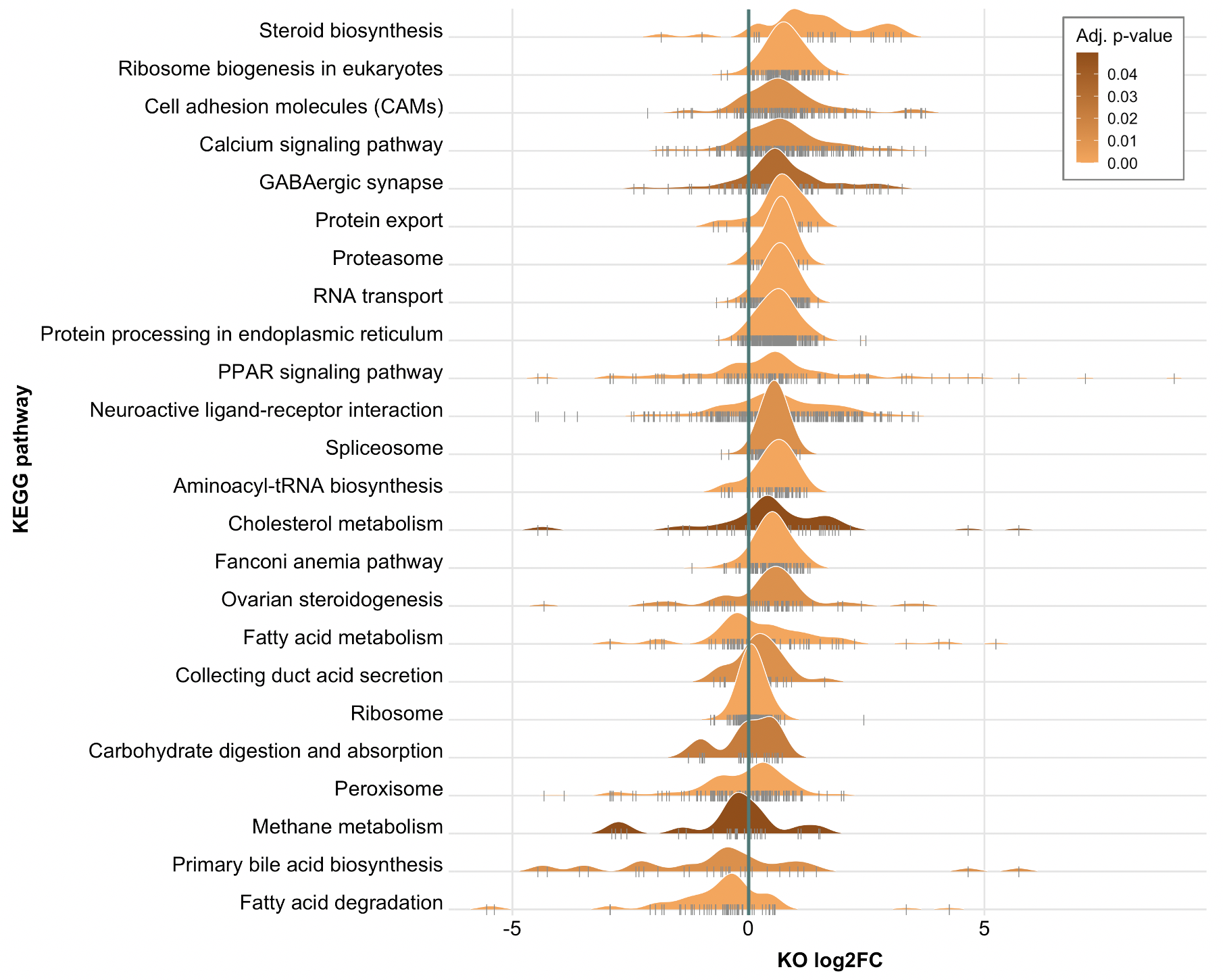
Significantly enriched pathways computed using GSEA for DCCO.
The absolute values of KO log2FCs were used for GSEA.
Pathways were considered significant if the adjusted p-value was < 0.05.
The log2FC of all genes in the enriched pathways are indicated with the vertical grey lines.
The distribution for each pathway is colored according the pathway’s adjusted p-value.
